Navigation auf uzh.ch
Navigation auf uzh.ch
-> see our guidelines on Github
A collaborative effort with INDEPTH members: Dumur, Duncan, Graumann, Desset, Randall, Mittelsten Scheid, Bass, Prodanov, Tatout & Baroux (2019) Nucleus, 10:1, 181-212

 Nuclear organization dynamics during cellular reprogramming
Nuclear organization dynamics during cellular reprogramming
The nucleus is more than a genetic container. This organelle is the chief orchestra of cellular processes by controlling and fine-tuning gene expression in response to developmental and environmental cues. We are interested in the 3D, functional organization of the genome in the plant nucleus. Our research aim to elucidate chromatin dynamics principles underlying cellular reprogramming during developmental or physiological transitions. Focusing on two case studies, the somatic-to-reproductive cell fate transition, and the dark-to-light physiological transition, we deploy a combination of genetic, molecular, cell biology and cytogenetics approaches to describe chromatin organization at a microscopic and nanoscopic scale. We established protocols for high-to-super resolution, 3D quantitative analyses of chromatin composition and organization at the single-cell level in whole-mount plant tissues or isolated nuclei.
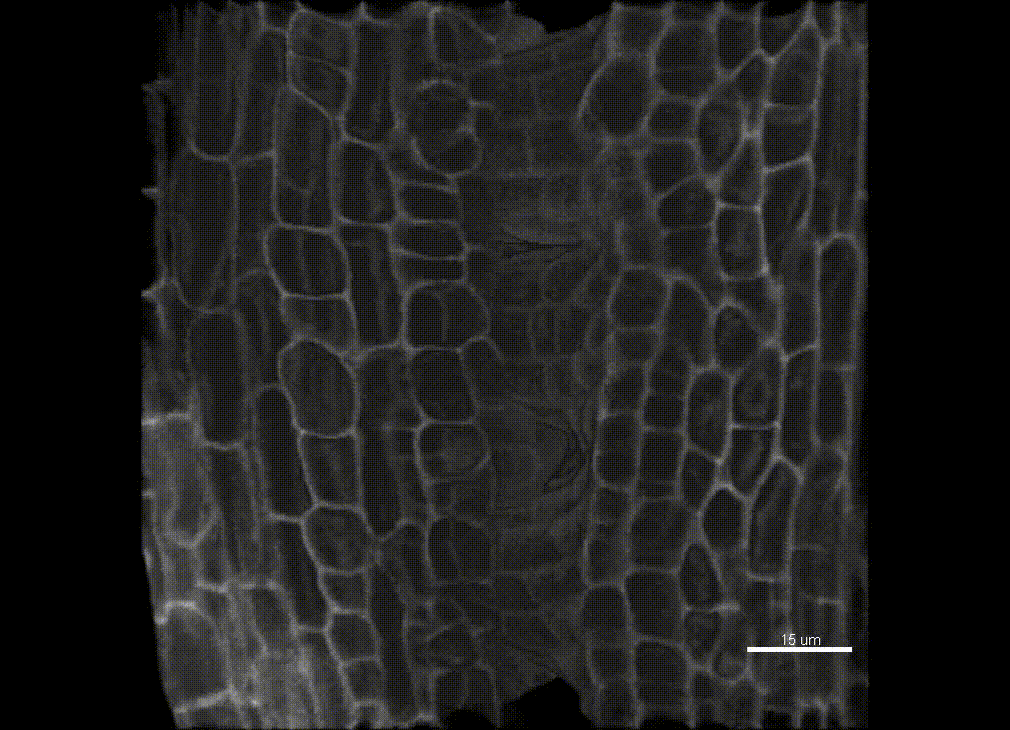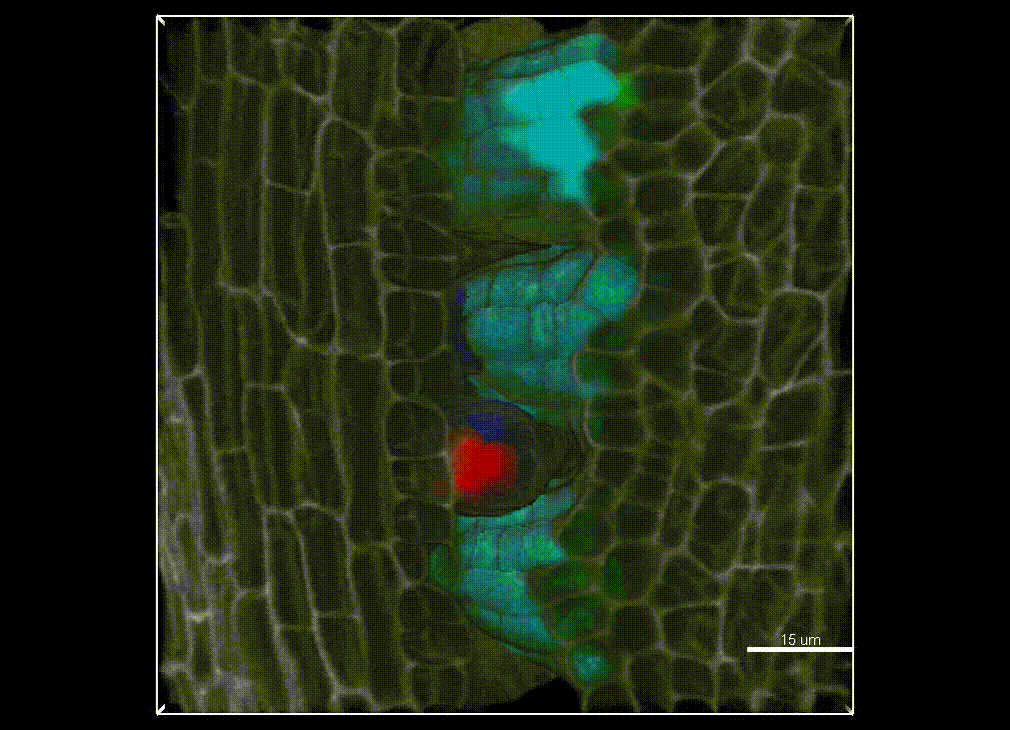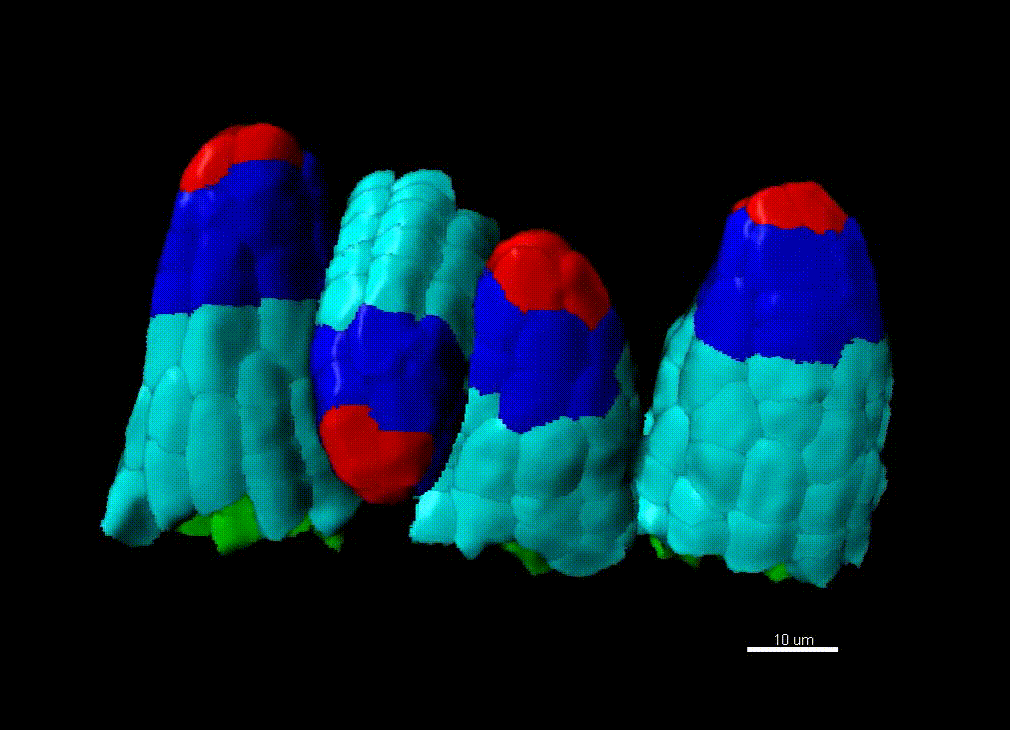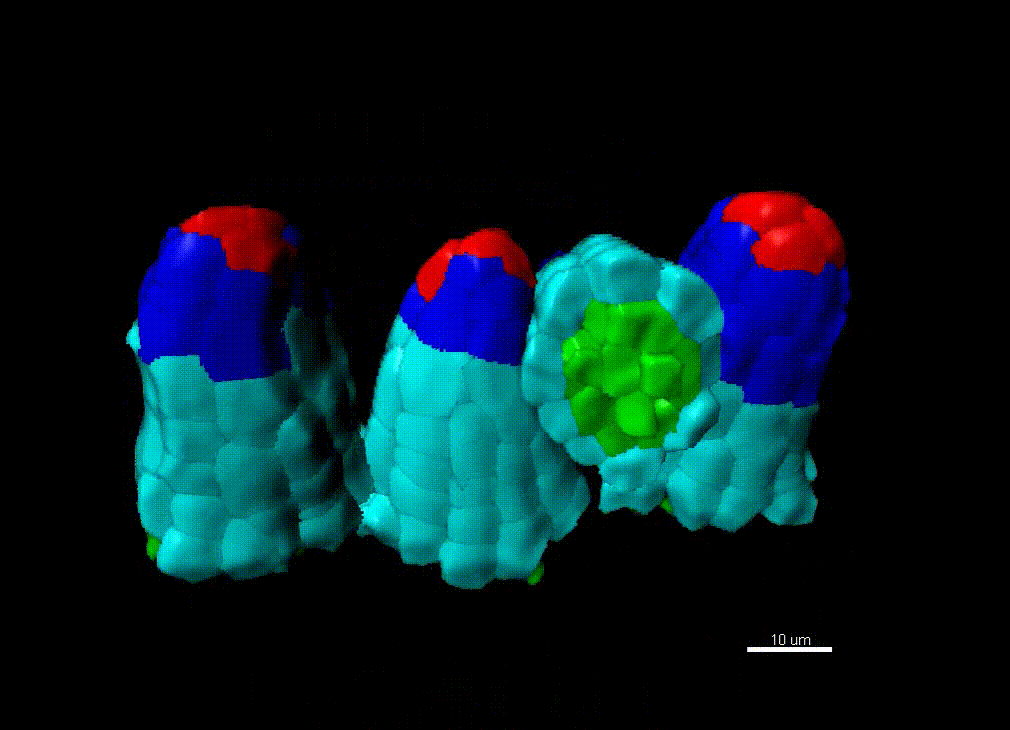 Contribution of organ growth to germline fate differentiation in the plant ovule
Contribution of organ growth to germline fate differentiation in the plant ovuleOrgan shape is contributed by coordinated tissue growth and cellular differentiation involving an interplay between mechanical constraints and gene regulation (Whitewoods and Coen 2017, Johnson and Lenhard 2011). We are interested in understanding the contribution of growth constraints in the plant ovule to the differentiation of the spore mother cells at the somatic-to-reproductive cell fate transition. We deploy high-resolution imaging and 3D image processing approaches to quantify tissue growth in 3D at the cellular level.
*************************************************************************************************
Support in past and present years
University of Zurich, Swiss National Science Foundation (SNSF), SystemsX.ch, Zurich-Basel Plant Science Center/Syngenta PhD program, Swiss Commission for Technology and Innovation (CTI/KTI), Baugarten Stiftung, Velux Stiftung, Ricola Stiftung.
*************************************************************************************************
Yanru Li, Danli Fei, Kinga Rutowicz, Célia Baroux
Former member: Jasmin Schubert
Funding: SNSF, University of Zürich
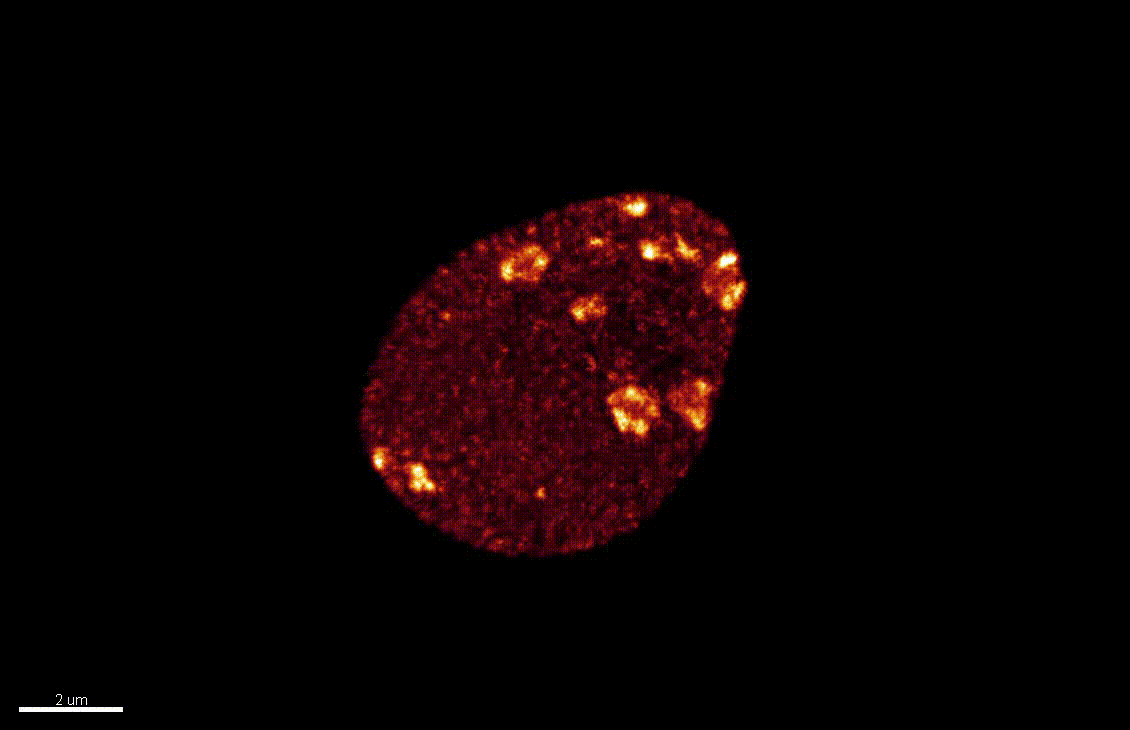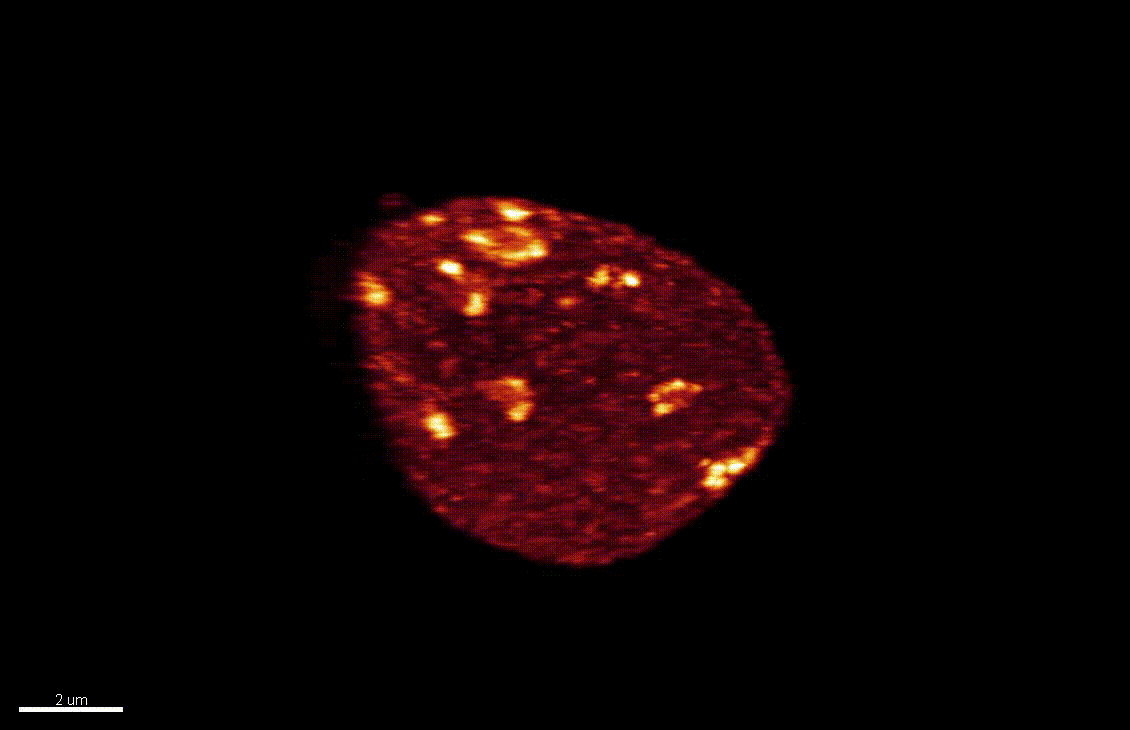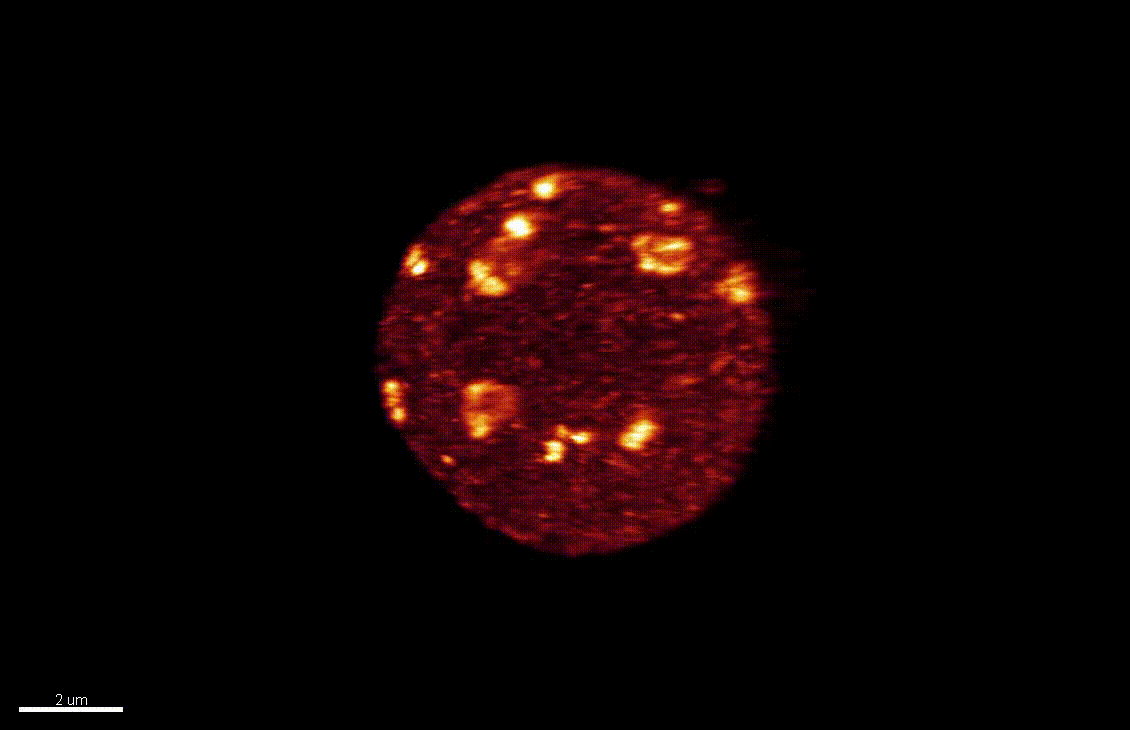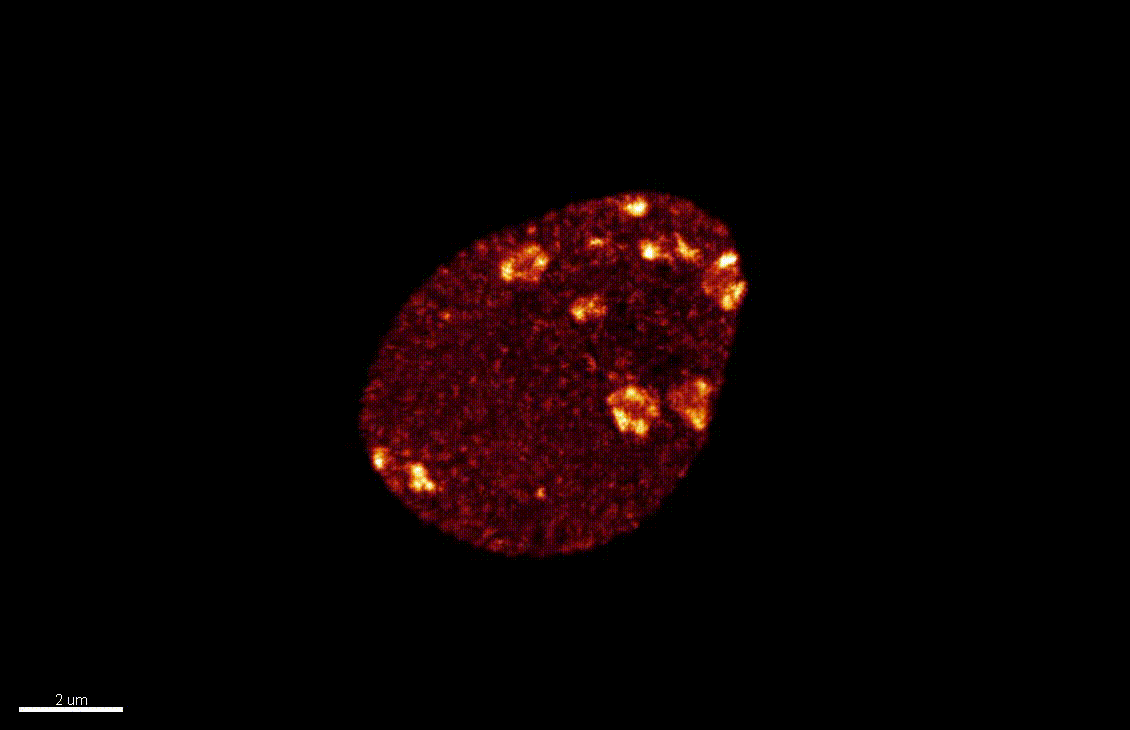
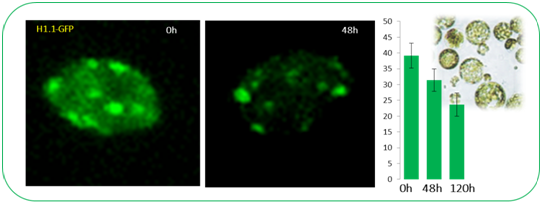
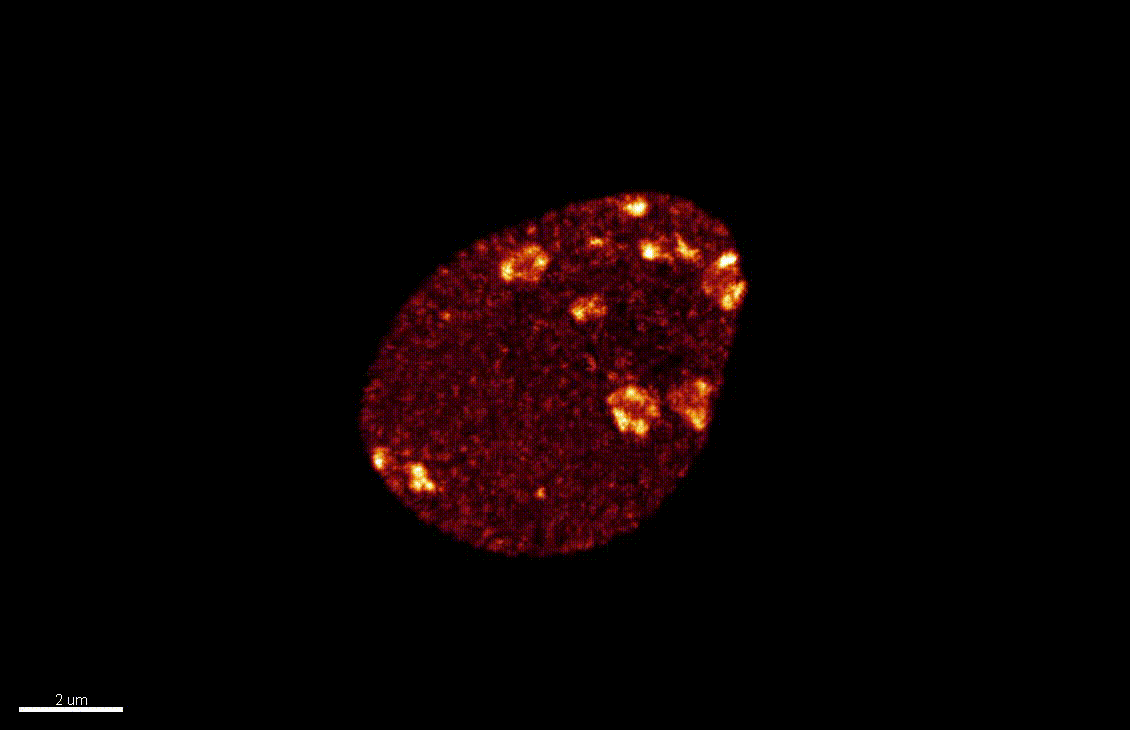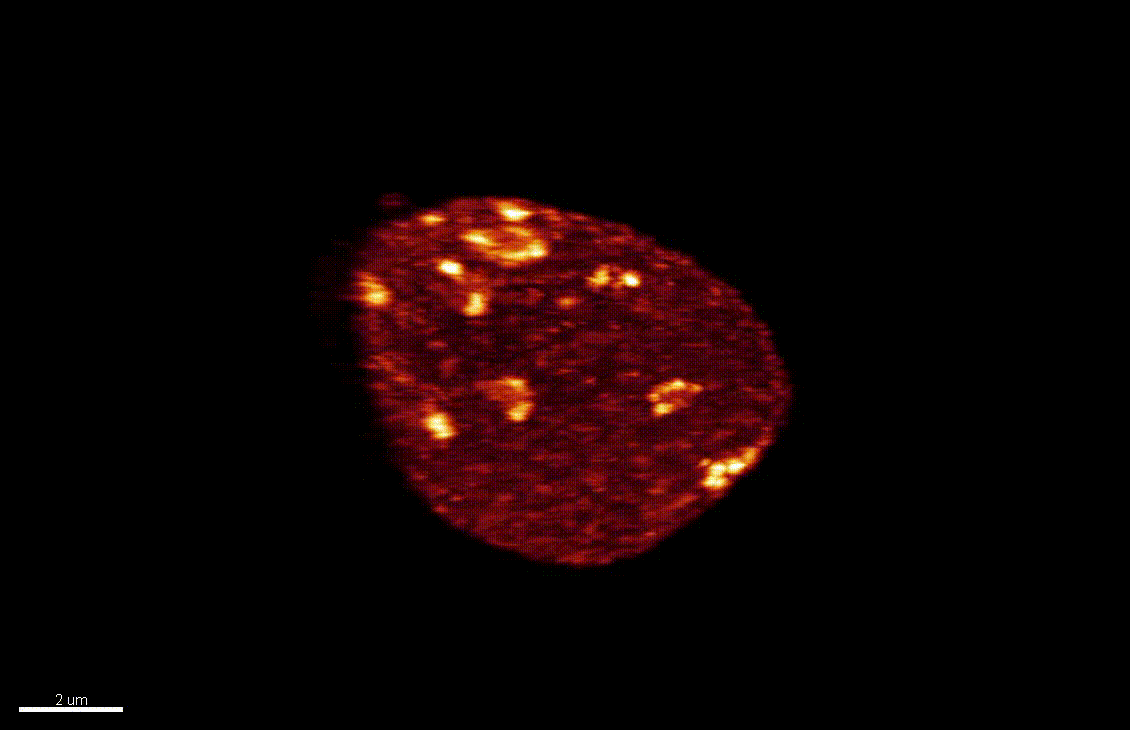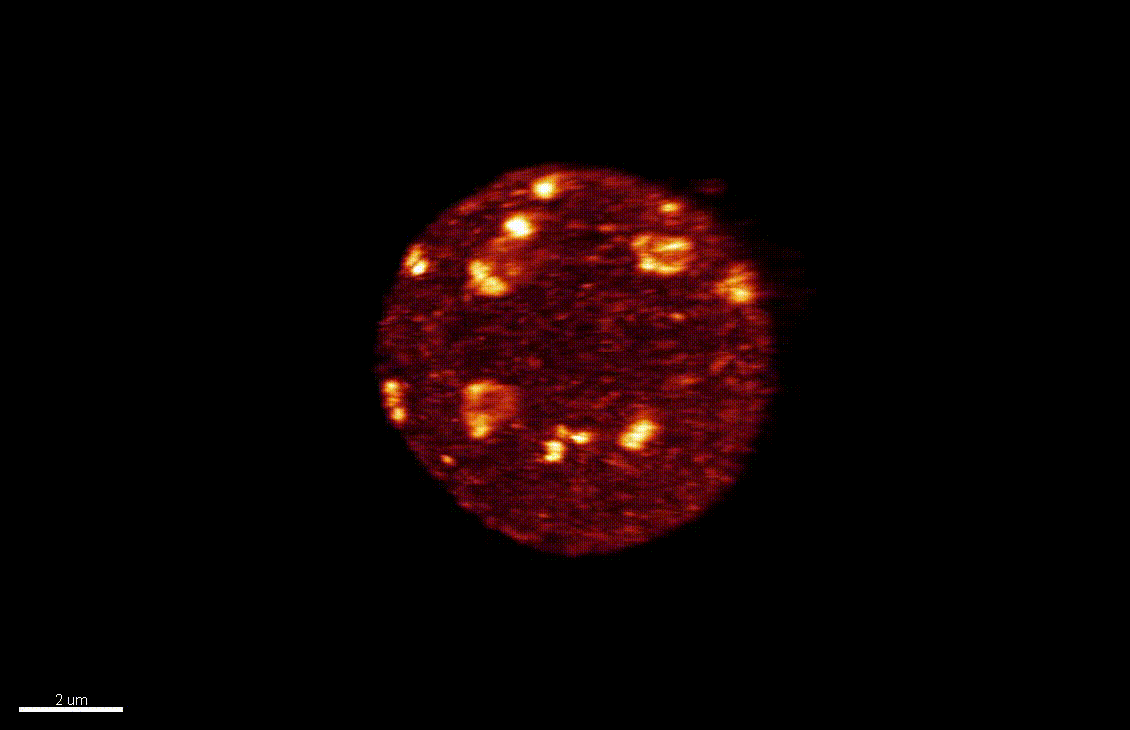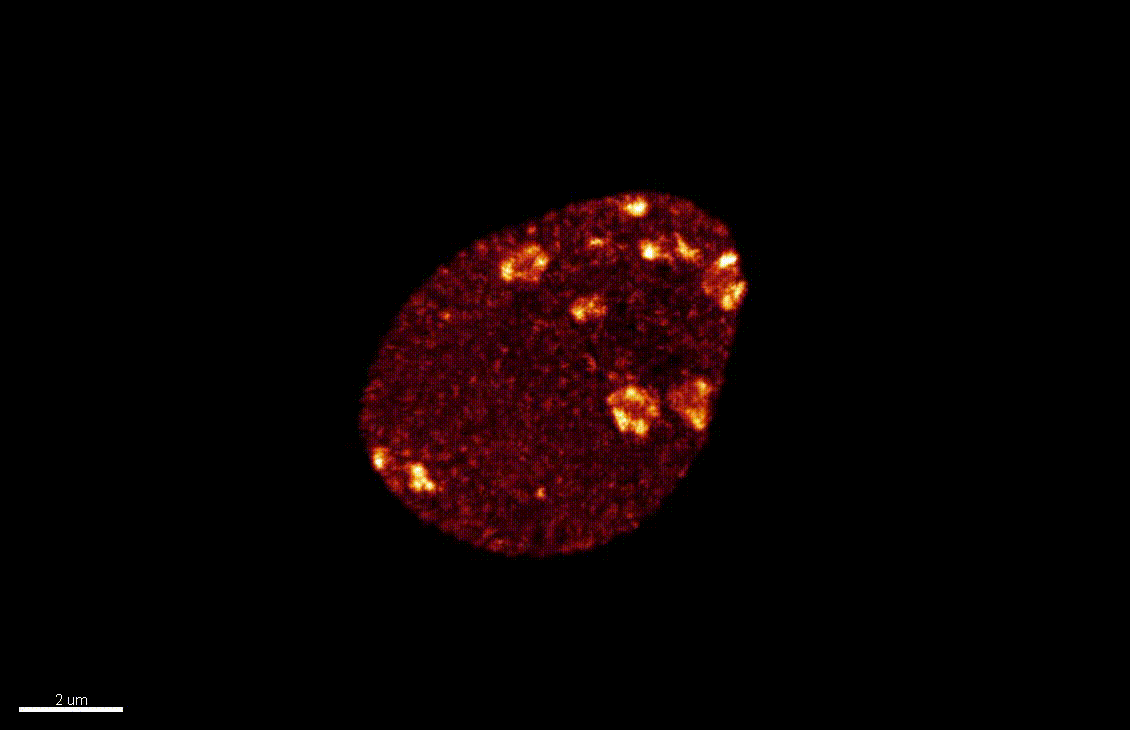
The differentiation of spore mother cells (SMCs) marks the somatic-to-reproductive fate transition in plants. Meiosis entails the formation of the haploid multicellular gametophytes, from which the gametes are derived, and during which epigenetic reprogramming takes place. We have shown that in the Arabidopsis both male and female SMC differentiation are accompanied by large-scale chromatin reprogramming characterized by chromatin decondensation, reduction in heterochromatin, depletion of linker histones, changes in core histone variants and in histone modification landscapes (She et al., 2013, She et al. 2015) Genetic analysis indicated that these events contribute to establishing postmeiotic competence, ie permissive o the development of the pluripotent gametophyte. In this study, we found that the eviction of linker histones H1.1 and H1.2 is a precocious event contributed a proteasome-mediated degradation. We are now focusing on elucidating the mechanisms of H1 dynamics by engineering conditional degradation-resistant versions and manipulating candidate chaperones. These genetic perturbation tools, together with developmental and molecular profiling analyses will allow elucidating the immediate and/or long-term function of H1 dynamics.
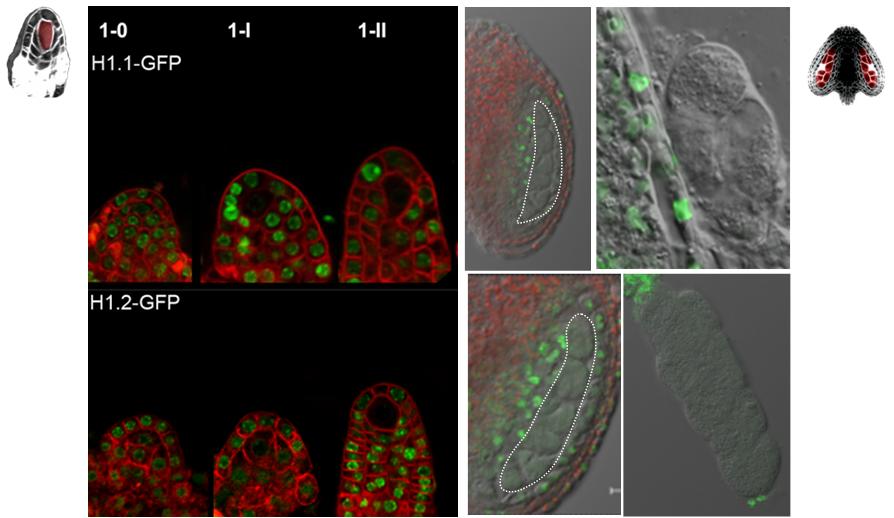
H1.1-GFP, H1.2-GFP (green), FM4-64 (left panel, red) or, chloroplasts (red, right panel).
From She et al. Development 2013, She and Baroux. Frontiers in Plant Sciences 2015.
Kinga Rutowicz, Célia Baroux
Assistant: Jerome Julmi (2019)
Collaborations: Prof. Lucas Pelkmans group (IMLS, University of Zurich). Prof. Andrzej Jermanowski (IPB, Warsaw, Poland)
Funding: Sciex project 13.229 (2014-2015), Plant Science Fellowship and Forschungskredit to KR (2018), SNSF (2019-2020)
Following the observation that H1’s are rapidly lost at the onset of cell fate transition in SMC differentiation (She et al 2013), we asked whether H1 eviction underlies other cellular reprogramming events. Cellular transdifferentiation upon protoplast release and culturing represents a suitable, well characterized system to follow and manipulate chromatin reprogramming. Arabidopsis protoplast cells derived from leaf tissue were shown to undergo drastic nuclear reorganization during trans-differentiation, including reduction in heterochromatin, dispersion of centromeric repeats, redistribution of histone modifications (Tessadori et al, J Cell Sci. 2007;120:1200-8). Consistently, we observed rapid a loss/reduction of H1-GFP signals in leaf protoplasts during the de-differentiation phase. We are undertaking a genetic screen, using conditional amiRNA-based silencing against candidate genes in a dual reporter background line to identify factors controlling H1 eviction and heterochromatin decondensation. Together with the group of Prof. Lukas Pelkmans, we are optimizing a semi-automated, microscopy-based phenotyping and nuclei classification workflow. In addition, we are characterising further the phenotype of H1-deficient plants in callus and shoot regeneration (Rutowicz et al 2019)
Fillipo Mirasole, Lena Perseus and Célia Baroux
Former member: Ricardo Randall
Collaborative project together with Dr Fredy Barneche group (IBENS, Paris, France) [Link]
Funding: Velux Stiftung, Ricola Stiftung
Project Website [Link]
Seedlings exposed to light for the first time, following germination, undergo a rapid reprogramming of gene expression. The lab of Dr Barneche showed that this event is contemporary to massive chromatin reorganisation (Bourbousse et al. 2015 PNAS 112(21):E2836-44) and found a candidate chromatin remodeler central to this process. In this collaborative project, we will elucidate the molecular and structural variations of the chromatin rapidly induced by light perception, aiming for unprecedented levels of details using state-of-art molecular profiling and super-resolution microscopy imaging.
Elizabeth Kracik-Dyer, Célia Baroux
Former member: Ricardo Randall
Collaboration: Dr. Stefan Grob, University of Zurich
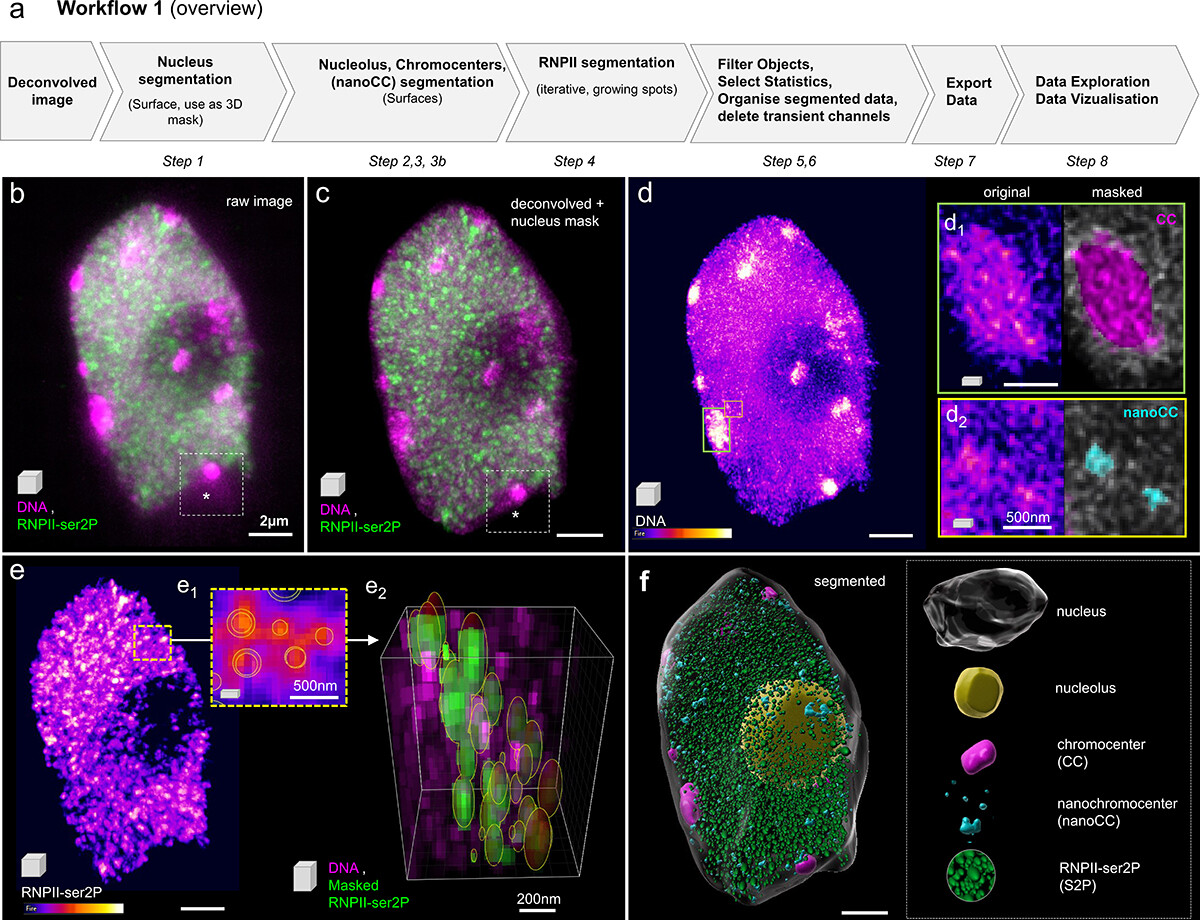
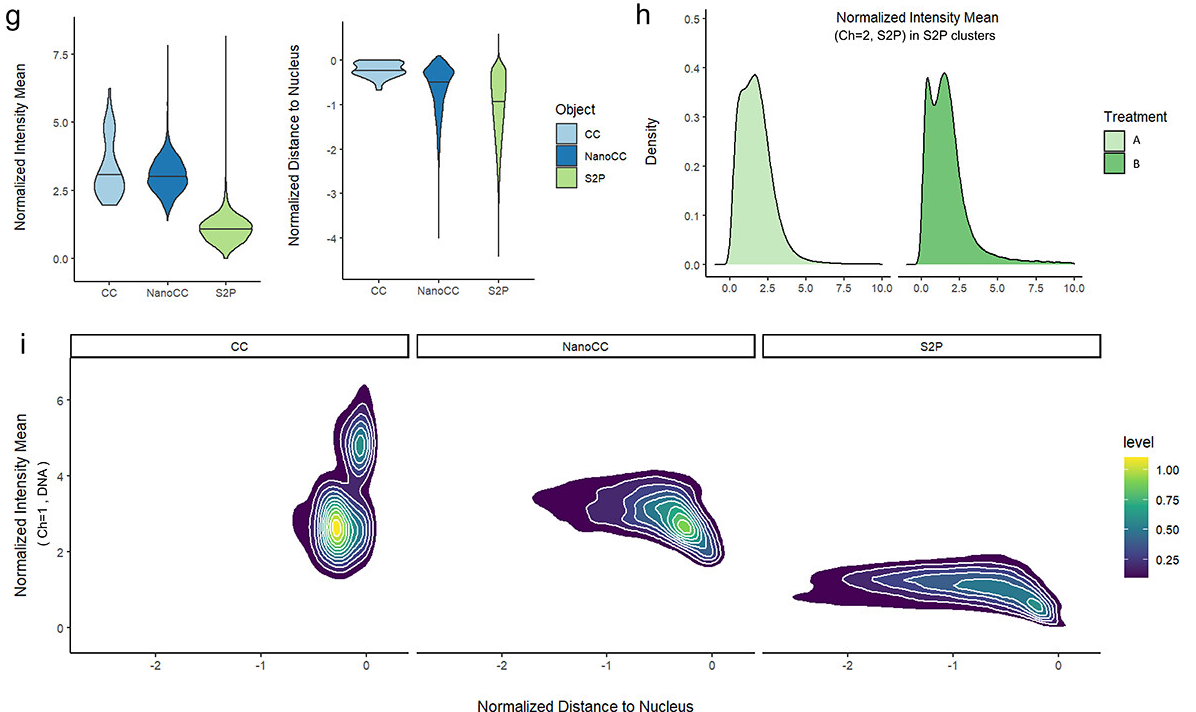
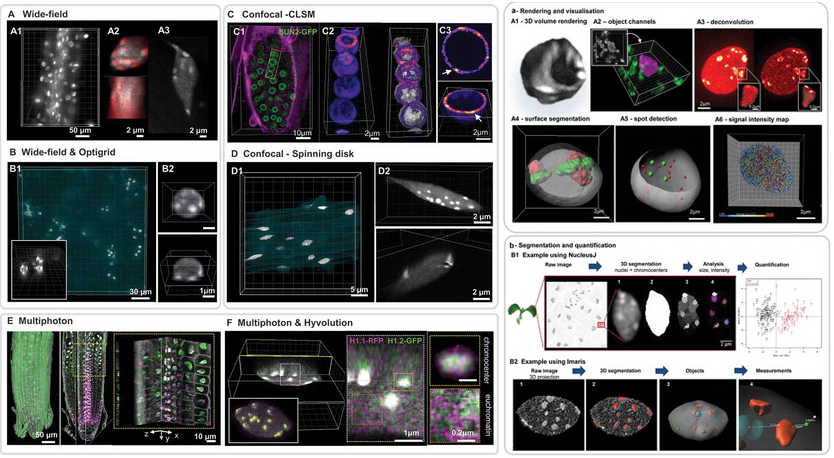
The nucleus is more than a genome-packaging organelle. Decades of studies, principally performed in yeast and animal cells, revealed key organizing principles of the interphase nucleus that influence nuclear functions. We are investigating the spatial organization of chromatin domains at the nanoscale and that of the transcriptional compartment in Arabidopsis. We use immunolabeling, 3D STED imaging, multiphoton imaging and quantitative image processing using customized Imaris workflows (Randall et al 2022 and Dumur et al 2019) and a custom data visualisation interface (DataViz and see Randall et al 2022)
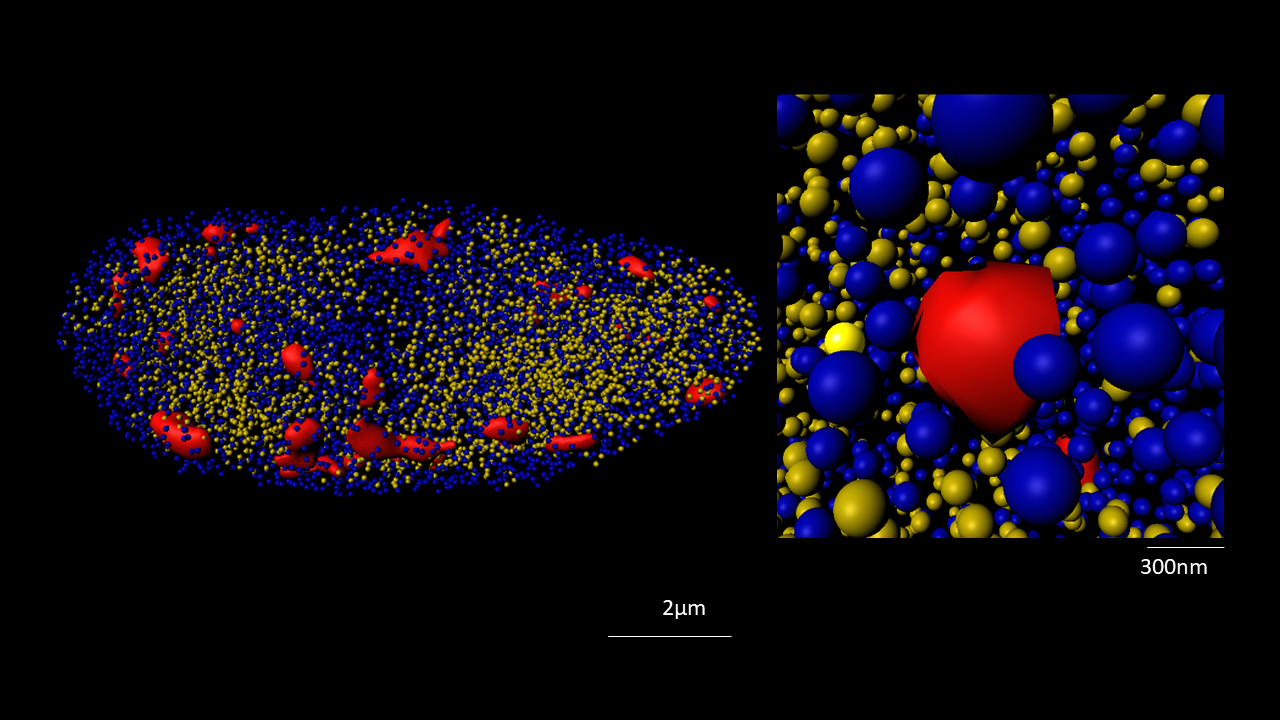
Ethel Mendocilla-Sato, Gabriella Mosca, Nuno Pires, Célia Baroux
Elvira Hernandez-Lagana, Daniel Grimanelli, Daphné Autran (IRD Montpellier, France)
IMAGO consortium: D. Autran, D. Grimanelli (IRD Montpellier, France); C. Godin (INRIA, France); O. Hamant, A. Bouaoud (ENS Lyon). Link
Funding: CTI (2014-2015), SNF/ANR Bilateral project (2016-2019), Forschungskredit to GM
Project closed
Publications:
In plants, the germline is established late in development. The differentiation of spore mother cells (SMC) is stereotypical in the Arabidopsis ovule: the female SMC differentiates in the upmost, central and subepidermic position of the digit-shaped ovule primordium. SMC differentiation and unicity is controlled by epigenetic and cell signaling mechanisms but is also intimately linked to ovule primordium growth. To elucidate the relationship between patterning and SMC establishment, we established a cell-based atlas of ovule primordium growth in wild-type and mutant lines. We use 3D CSLM and LSM imaging followed by cell-based segmentation (link to our protocol), quantitative analyses of growth per domain and cell layer, reporter and genetic analyses
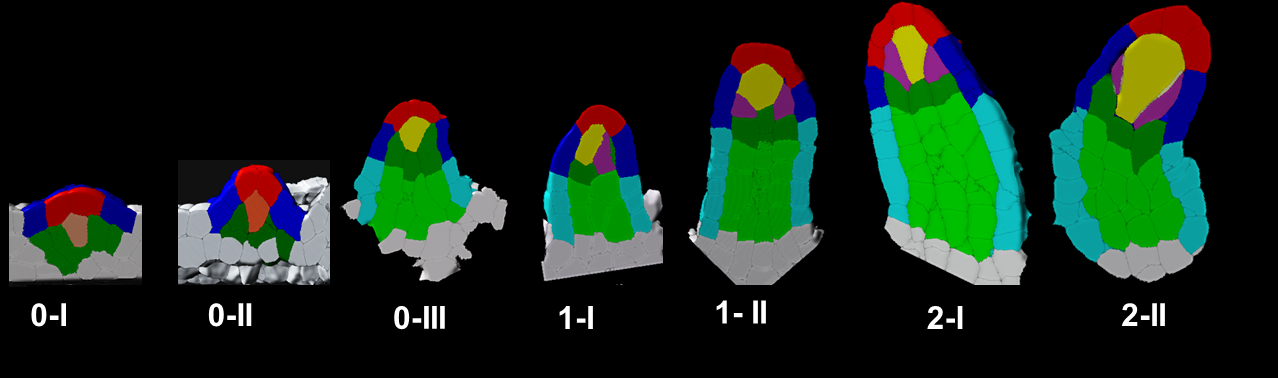
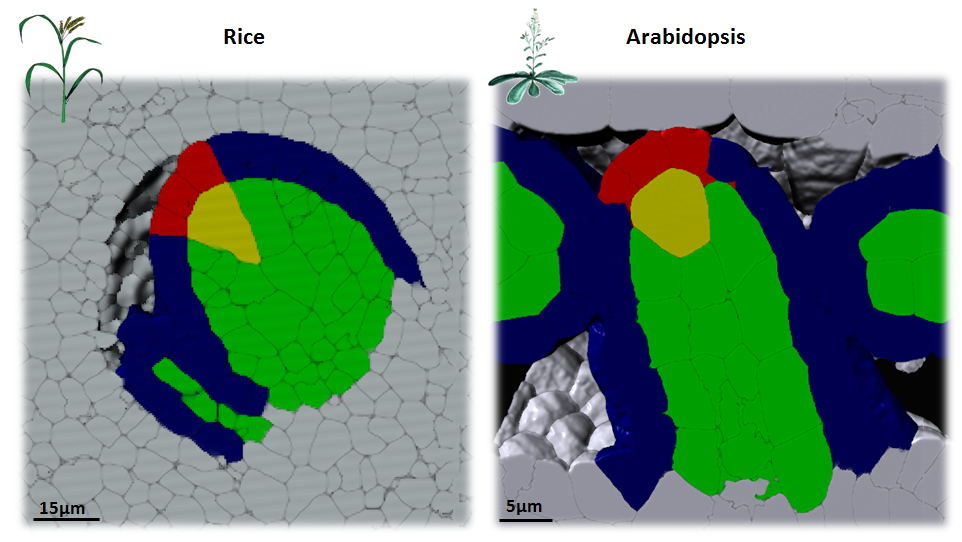
Célia Baroux, Stefan Wyder, Ueli Grossniklaus, Jordi Romero, Claudia Köhler
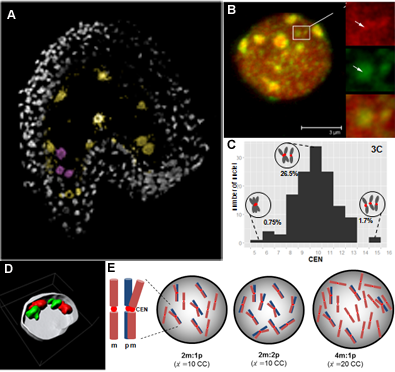
Reproductive success relies on the endosperm, an extra-embryonic nurse tissue produced at fertilization. The dosage of paternally vs maternally-derived products defines a fine balance critically influencing its development, where alteration can result in seed abortion and hybridization failure (reviewed in Lafon-Placette and Köhler, Mol Ecol 25, 2016). We discovered a specific heterochromatin fraction (ESI) which responds to ploidy alterations in a parent-of-origin-dependent manner (Baroux et al., 2007) as well as to interspecific hybridization (Baroux, unpublished). One aim is to elucidate the mechanisms and targets of maternal ESI heterochromatin in relation to dosage regulation of parental genomes’ expression in the endosperm. Another aim is to determine genetic loci associated with ESI and possibly responding to parental dosage. We indeed identified a handful of genes enriched in a histone mark enriched at ESI foci and with a maternally-compensated gene expression (Romero, Wyder, Köhler, Baroux, unpublished). Whether those also relate to parental chromosome associations in the triploid endosperm (Baroux et al 2016) remains to be determined.
Kinga Rutowicz, Marek Whitehead, Célia Baroux
Collaborations: Lukasz Knizewski, Krzysztof Ginalski, Prof. Andrzej Jermanowski (IBB, University of Warsaw, Poland)
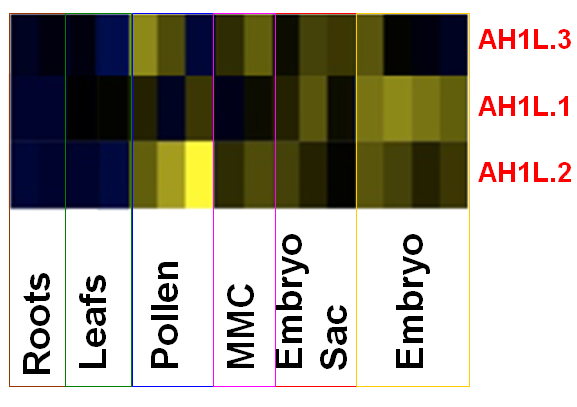
The reproductive lineage is largely devoid of the canonical H1s (with the exception of the transient, meiotic chromosomes). This raises the possibility that plants possess germline-specific H1 variants that may enable a relaxed chromatin structure favorable to reprogramming in the germline. By analyzing protein variants sharing homology with the globular domain of H1’s (GH1), we identified three atypical variants (AH1L.1-.3, Knizewski, Ginalski Jermanowski, unpublished) that show transcript enrichment in the SMCs, gametes and embryo according to published cell-specific transcriptome profiles (Schmid et al, PLoS Genetics 2011). We are characterizing their spatial and temporal expression profile using reporter lines, molecular and developmental function using a combination of genetic, cell and molecular approaches.