Navigation auf uzh.ch
Navigation auf uzh.ch
Mechanisms of genome evolution
Plant genomes are very dynamic and evolve much more rapidly than animal genomes. We are investigating the molecular mechanisms that drive this rapid evolution. Plants show an enormous range in genome size. For example, the genome of the Lily Fritillaria assyriaca is more than 1,000 larger than the genome of Arabidopsis. Genomes are expanded by the amplification of transposable elements (TEs), small genetic units which can move around in the genome and make copies of themselves. The expansion of genomes is counteracted by deletion of DNA. We have identified several ways in which DNA can be removed from the genome. A central mechanism is the repair of double-strand breaks (DSBs) which can be highly inaccurate in plants, leading to extensive deletions and rearrangements. It turns out that the activity of TEs is a major cause of DSB and therefore a major driving force in the evolution of plant genomes.
Comparative genomics
Comparative genomics focuses on the large-scale comparison of entire genomes. Typically, we compare gene content and gene order of two or more organisms. Comparative analysis is a powerful method to discover mechanisms that drive the evolution of genomes. We have so discovered multiple mechanisms that can lead to gene duplications, gene movement and gene loss. Of central importance to genome rearrangements is double-strand break (DSB) repair. DSBs can be introduced by the activity of transposable elements. In plants, DSB repair is often error-prone, leading to the deletion, translocation and duplication of large genomic segments.
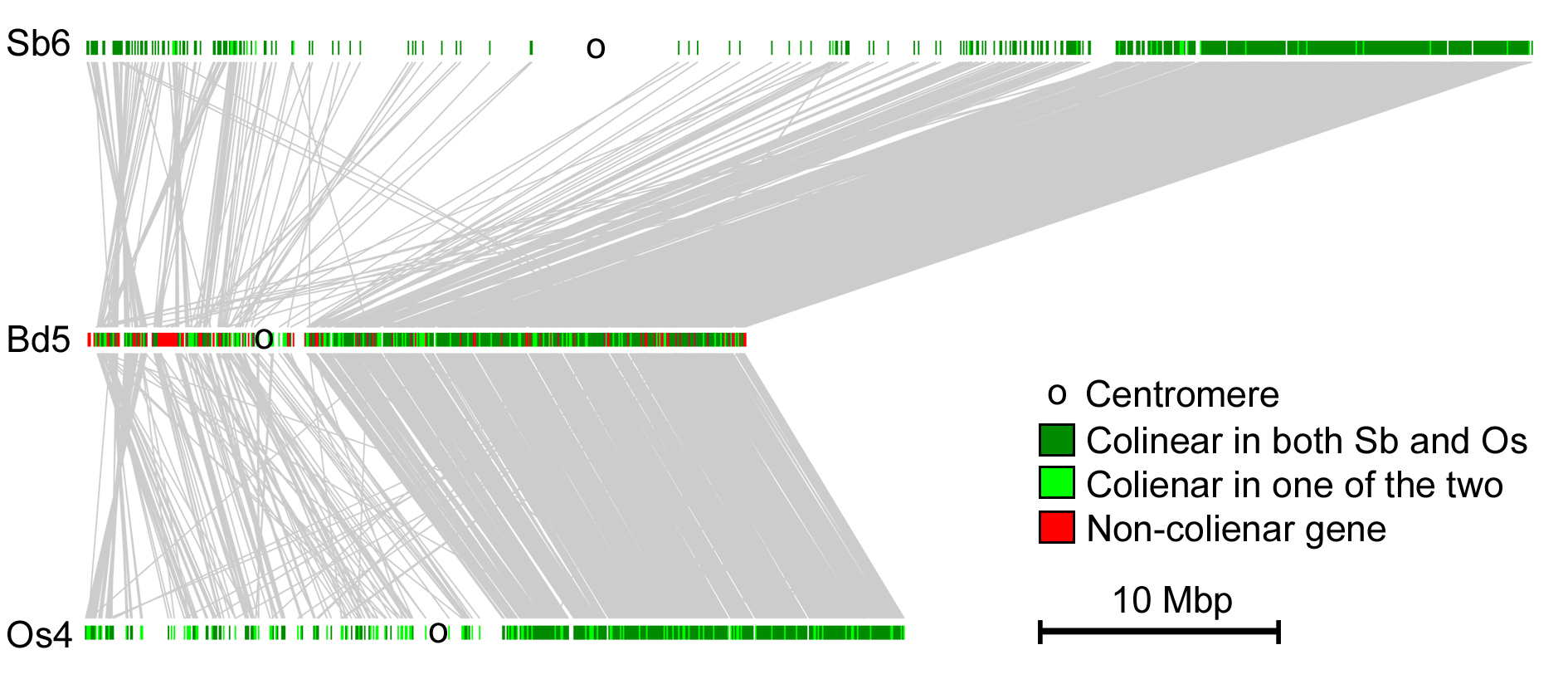
Comparative analysis of gene order and conservation in grass chromosomes. Grass show a strong conservation of gene order across most chromosomes. Here, chromosomes 6 from Sorghum bicolor (Sb), Brachypodium distachyon (Bd) and rice (Os) are compared. The centromeres are indicated by black circles. homologous genes are connected by gray lines and shown in green. In this case, gene order on the short chromosome arms is much less conserved than in the long arms. The reason for it is still unknown.
Evolution of interaction
Using comparative genomics and transcriptomics approaches, we are studying the interactions of plants and microbes. This includes plant/pathogen interactions as well as symbioses. In recent years we were involved in sequencing and analysis of the genome of Blumeria graminis, the cause of the wheat powdery mildew disease. We are studying genetic diversity of the pathogen as well as the response of the plant to pathogen attach. Additionally, we are studying symbiotic systems such as lichens (a symbiosis of fungi and algae) and the symbiosis of Psychotria plants with Burkholderia.
Development of Bioinformatics methods
The ever-increasing capacity of sequencing technologies poses an enormous challenge to biologists, since much more data is being produced than can reasonably be analysed. An important focus of our work is therefore to develop bioinformatics tools that allow complex analyses on very large datasets. We have a large collection of tools for whole-genome TE analysis, comparative genomics and data visualization. For more information on artistic visualization, visit the WICKERsoft homepage.
In 2020, we contributed to a study on hygiene in kitchen and common living spaces ("Küchenluft, Hygiene und Energie", KLEH) which was funded by a range of public and private institutions. We collected mircobial probes and sequenced and studied bacterial and fungal genomes. The full results are shown here.