Navigation auf uzh.ch
Navigation auf uzh.ch
Funding: SNF project 31003A_179322 (2019-2023)
Our research aims to answer the following fundamental questions:
People involved: Yilei Liu, Paula Bellés Sancho, Daphné Golaz
Collaborators: Dr. Christian Ahrens (Agroscope Wädenswil, Swiss Institute of Bioinformatics), Prof. Dr. Nicola Zamboni (IMSB, ETHZ).
To understand the molecular processes underlying β-rhizobial symbiosis we are applying a multi-layered functional genomics analysis on root nodules and free-living rhizobia by comparative genomics, RNA-Seq, shotgun proteomics and metabolomics (Figure 1). These state-of-the-art technologies combined with classical phenotypical approaches allow us to identify 1) β-rhizobial genes/proteins important for symbiosis and 2) mechanistic differences between α- and ß-rhizobial symbiosis. After 50 million years of separate evolution we expect that α- vs. β-rhizobial symbiosis should be very different. We use the symbiosis between B. phymatum STM815 and the legume Phaseolus vulgaris (bean) as a model.
Integration of transcriptomics, proteomics and metabolomics datasets (Figure 1) derived from the global studies followed by a careful bioinformatic analysis through classification of genes in functional categories, assignment to pathways, gene-set enrichment analysis (GSEA) and literature searches are used to prioritize Paraburkholderia genes specifically expressed during symbiosis and potentially important for symbiosis with legumes. Mutants in genes of interest are generated and characterized in vitro for growth, production of secondary metabolites, resistance to environmental stress and in vivo for the capability to efficiently enter symbiosis with legumes (Figure 2). We recently found that - in contrast to most rhizobia - Paraburkholderia strains can synthetize the essential component of the nitrogenase enzyme, homocitrate, through NifV (homocitrate synthase) and that NifV plays a key role for nitrogenase activity during symbiosis and in free-living conditions (Bellés-Sancho et al 2021a).
Moreover, metabolomics and dual RNA-sequencing on root nodules revealed that the master regulator of nitrogen fixation NifA is also involved in controlling other cellular functions such as auxin production and stress response (Bellés-Sancho et al 2021b).
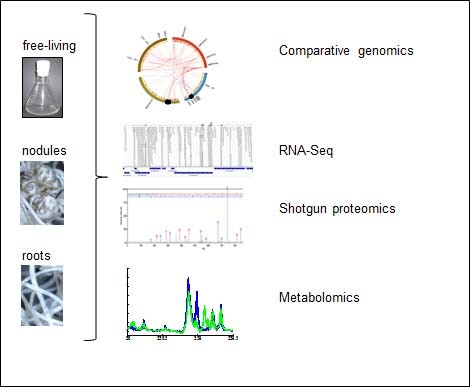
Figure 1. Functional genomics on P. phymatum living in culture (in vitro) and in Phaseolus vulgaris root nodules and roots (in vivo).
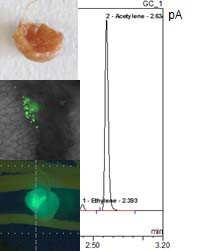
Figure 2. Morphology of P. vulgaris root nodules containing P. phymatum STM815 wild-type strain. A confocal microscope is used to investigate a root nodule of a P. vulgaris plant infected with a gfp-tagged B. phymatum strain (40x magnification). The nitrogenase activity of root nodules is measured using the acetylene (C2H2) reduction assay. The presence of an ethylene peak (C2H4) shows that the P. phymatum nodules are synthetizing active nitrogenase.
People involved: Paula Bellés Sancho, Sebastian Hug, Yilei Liu
Collaborators: Prof. Hans-Martin Fischer (ETHZ), Dr. Lionel Moulin (IRD, Monpellier)
Several legume plants can be efficiently nodulated by alpha- and beta-rhizobia. To answer the question whether alpha- and beta-rhizobia are able to compete for nodule occupancy, we are performing co-inoculation experiments using different alpha- and beta-rhizobial strains and different legumes under different environmental conditions. To this end, differentially tagged strains are used in competition assays. This will allow to follow the interaction and to determine the distribution and proportion of alpha- and beta- rhizobial strains inside the root nodules using a confocal laser microscope (Leica TCS SPE). We showed that Paraburkholderia phymatum is highly competitive against other beta-rhizobial strains for nodulating several papilionoid legumes of major agricultural importance including the common bean (Lardi et al 2017).
Competition for root infection has been shown to depend on environmental conditions, geographical position and on symbiont and host plant genetic diversity. In our lab we focus on identifying the genetic traits important for nodulation competitiveness of beta-rhizobia with a special interest on bacterial secretion systems (de Campos SB et al 2017)(Hug et al 2021) and antibiotic production as well as exopolysaccharide production (Liu et al 2020).
Figure 3. Anti-bacterial activity by A) type-VI secretion systems (T6SS) and B) bacteriocin production