Navigation auf uzh.ch
Navigation auf uzh.ch
Our work is focused on the functional roles of three-dimensional chromosomal architecture. We are using state-of-the-art molecular biology tools, such as Hi-C and classic approaches, such genetics to show that 3D-chromosome folding is much more than simple packing of long chromosomes into tiny nuclei.
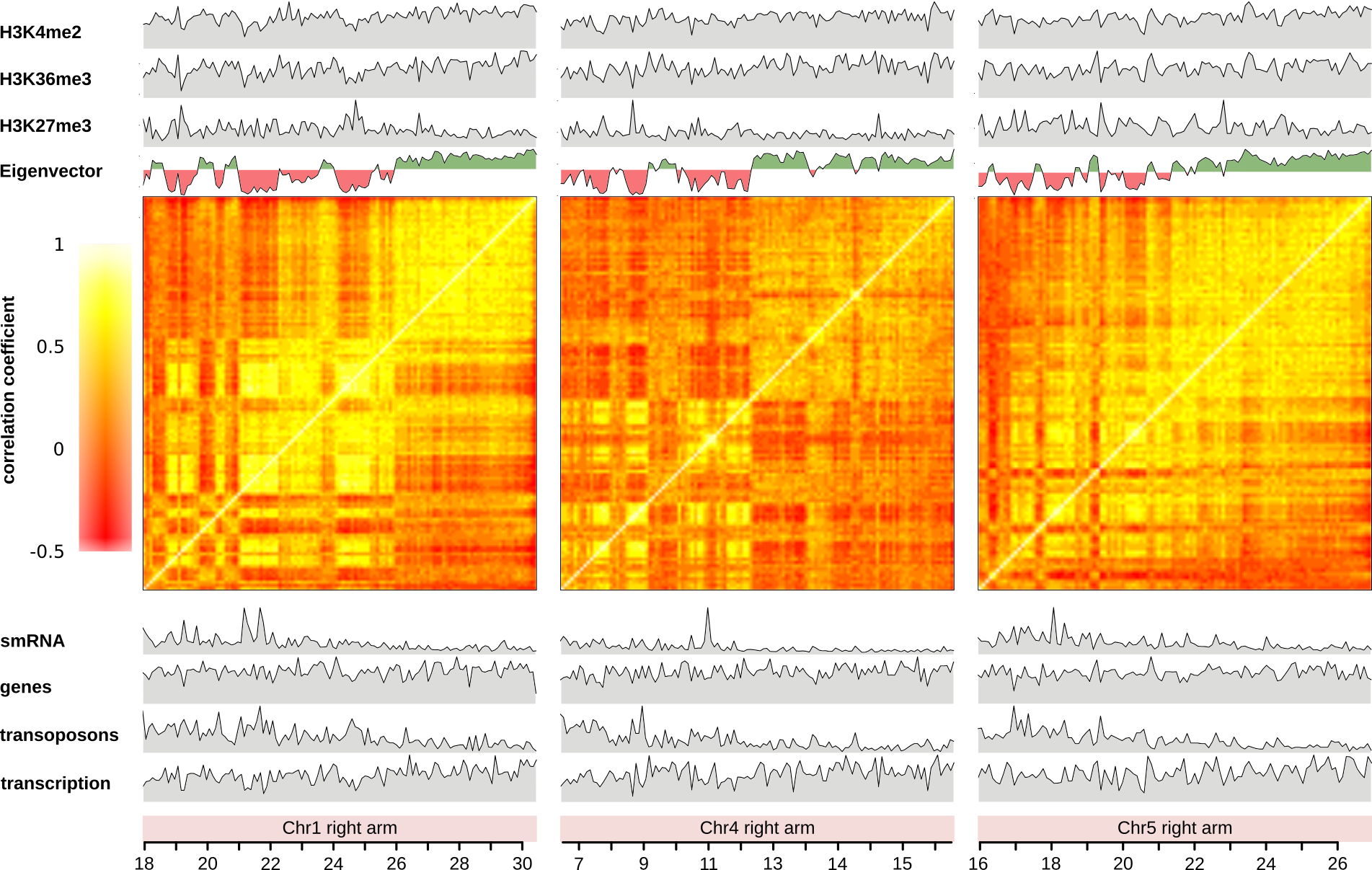
Over many decades, genomes and genome evolution have been predominantly studied in terms of the function of the therein-encoded genes. With the emergence of high-throughput DNA sequencing technologies, we gained a more holistic view on genomes and learned that they are highly organized and regulated structures. In addition to the linear genome, the epigenetic landscape, subdivides entire chromosomes in several regions with distinct epigenetic signatures. The epigenetic landscape is shaped by chemical modification of chromatin (the unity of both DNA and associated proteins). It concerns both, the DNA (e.g. methylation of cytosine) and associated proteins (e.g. methylation/acetylation of histones). These epigenetic signatures are important regulators of the activity of the genes and other genomic features. Thus, not only the DNA code determines genome function, but also additional levels of genome organization have to be taken into account when genome function is studied. Today, in addition to the DNA sequence and the epigenetic landscape, we are aware of a third organizational level, which previously did not receive sufficient attention, namely the 3D-genome organization.
We are interested in the interplay of the different organizational levels of the genome. Thereby, we especially focus on the relationships that 3D-genome organization has with both, the DNA sequence and the epigenome. The study of the influence of the DNA sequence (genetics) and the epigenome (epigenetics) on both, immediate cellular functions and evolution have revolutionized biological research. We are intrigued by the possibility that 3D-chromatin folding may also contain information and, hence, may exert an important biological function in both, micro-scale cellular functions and, macro-scale evolution.
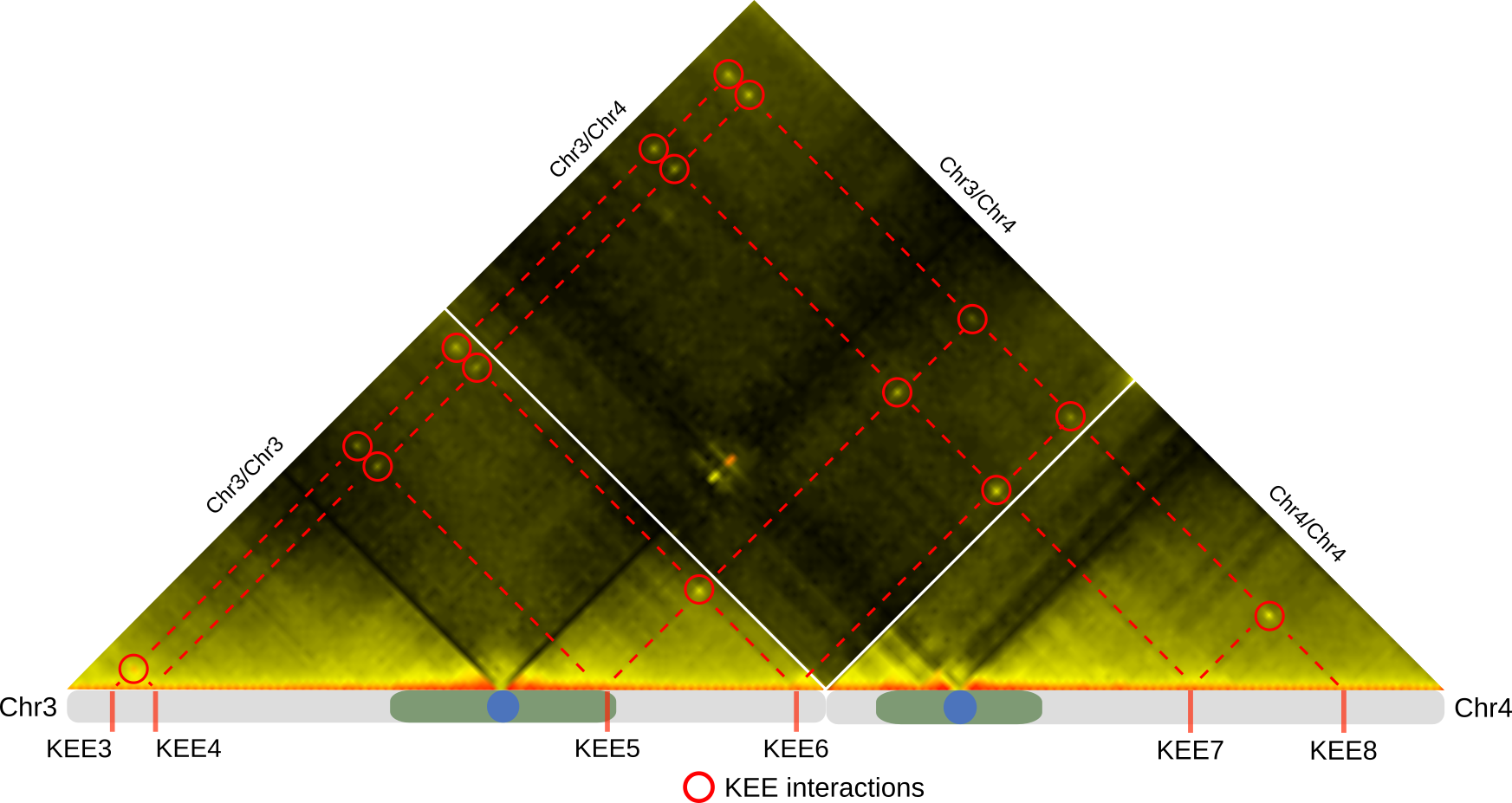
As for micro-scale processes, we could show that a specific 3D-chromatin structure, termed the KNOT is involved in the genome’s immune system. The KNOT is formed by tight 3D association of ten KNOT Entangled Elements (KEEs), which are enriched in both transposable elements and associated small RNAs. Invasive DNA elements, such as transgenes can be associated in 3D with the KNOT, which eventually leads to complete shutdown of these parasitic DNA elements.
Using molecular and -omics approaches, I am currently trying to understand the underlying mechanism of KNOT-linked silencing. Additionally, we initiated projects, which try to unravel the importance of 3D-genome architecture on genome evolution.