Navigation auf uzh.ch
Navigation auf uzh.ch
As a way to tackle the antibiotic resistance crisis natural killers of bacteria like predatory bacteria have gained attention in the scientific community recently1. Gram-negative predatory bacterium B. bacteriovorus has not only a big potential as a putative ‘living antibiotic’ by invading a wide range of Gram-negative (antibiotic resistant) pathogens in/on different organisms2-8, but as well as a biotechnological tool9 or as a source of potential new drugs due to its predatory and lytic capacity towards Gram-negative bacteria. B. bacteriovorus is ubiquitously present in the environment10-18 and a model organism for predatory bacteria that invade and replicate in the periplasm of other Gram-negative prey bacteria (Fig. 1)19. Once nutrients become scarce B. bacteriovorus septates to produce progeny which escape the depleted host cell remnants in the exit phase20 (Fig. 1, highlighted in yellow).
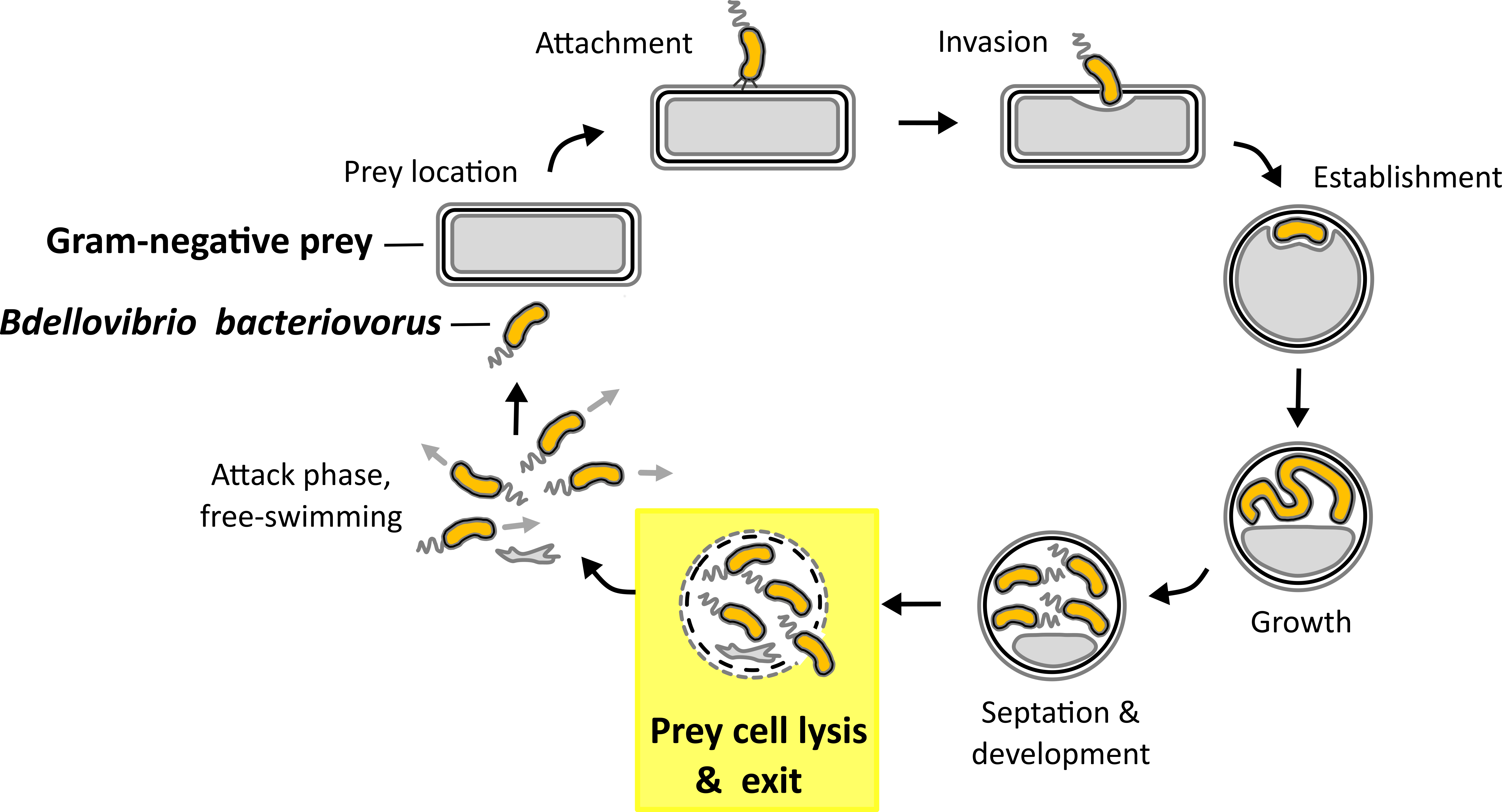
Fig. 1. Predatory life cycle of B. bacteriovorus HD100. In the predatory life cycle predator B. bacteriovorus divides within the periplasm of the Gram-negative prey bacterium. In the exit phase juvenile prey cells leave the depleted prey cell.
To date the molecular mechanisms underlying this last, crucial phase remain enigmatic apart from a novel predatory lysozyme that Simona characterized in collaboration with Prof. R. E. Sockett (University of Nottingham, UK) & Dr. A. L. Lovering (University of Birmingham, UK) during her SNSF Postdoc.Mobility fellowships21. This predatory lysozyme specifically recognizes and cleaves the nonself-marked (deacetylated GlcNAc) peptidoglycan in the prey bacterial cell wall as part of the prey exit and is potentially useful as a drug21. However, the prey exit mechanism involves degradation of other cellular structures with new predatory mechanisms to discover. We aim to learn from predatory bacteria with the help of state-of-the-art ‘omics’ technologies (transcriptomics, proteomics, metabolomics) and down to the detailed level of predatory molecular mechanisms. This knowledge is essential in order to use predatory bacteria or their novel enzymes in potential applications as biotechnological tools or therapeutic agents/drugs.
The project 'Learning from predatory bacteria: from 'OMICS' to molecular mechanisms' is funded by a Swiss National Science Foundation (SNSF) Ambizione Fellowship.
Video: Time-lapse microscopy of B. bacteriovorus
exiting of prey cell remnants of Escherichia coli. (Video by S. Huwiler)
Funding for other research projects on predatory bacteria was provided by:
- FreeNovation Grant (by Novartis Forschungsstiftung)
References:
1 Tyson, J. et al. Trends Microbiol 25, 90-91 (2017).
2 Sockett, R. E. et al. Nat Rev Microbiol 2, 669-675 (2004).
3 Negus, D. et al. Annu Rev Microbiol 71, 441-457 (2017).
4 Atterbury, R. J. et al. Appl Environ Microbiol 77, 5794-5803 (2011).
5 Scherff, R. H. Phytopathology 63, 400-402 (1973).
6 Cao, H. et al. Vet Microbiol 154, 413-418 (2012).
7 Dashiff, A. et al. J Appl Microbiol 110, 431-444 (2011).
8 Van Essche, M. et al. Mol Oral Microbiol 26, 52-61 (2011).
9 Martinez, V. et al. Sci Rep 6, 24381 (2016).
10 Stolp, H. et al. A van Leeuw J Microb 29, 217-248 (1963).
11 Jurkevitch, E. et al. Appl Environ Microbiol 66, 2365-2371 (2000).
12 Johnke, J. et al. Microb Ecol 79, 252-257 (2020).
13 Iebba, V. et al. PLoS One 8, e61608 (2013).
14 Fry, J. C. et al. Appl Environ Microbiol 31, 469-474 (1976).
15 Hobley, L. et al. BMC Genomics 13, 670 (2012).
16 Feng, S. et al. Environ Microbiol 18, 3923-3931 (2016).
17 Paix, B. et al. Appl Environ Microbiol 85, e02494-18 (2019).
18 Ezzedine, J. A. et al. Front Microbiol 11, 98 (2020).
19 Lambert, C. et al. Curr Opin Microbiol 9, 639-644 (2006).
20 Fenton, A. K. et al. J Bacteriol 192, 6329-6335 (2010).
21 Harding, C. J. et al. Nat Commun 11, 4817 (2020).