Projects
First quantitative proteome of periplasmic predation reveals a prey damaging protease
The rise of antimicrobial resistant pathogens calls for novel ways to kill and damage bacteria. A rich source for bacterial cell-damaging proteins is periplasmic predatory bacterium Bdellovibrio bacteriovorus, which invades, kills and subsequently exits the Gram-negative prey cell. An increased understanding of predatory protein function can be achieved by analyzing their relative abundance at key stages of predation. We present the first quantitative proteome covering the complete predatory life cycle of the bacterial predator B. bacteriovorus killing Escherichia coli, quantifying 2195 predator proteins. From these proteins, nine protein clusters sharing similar expression patterns were identified.
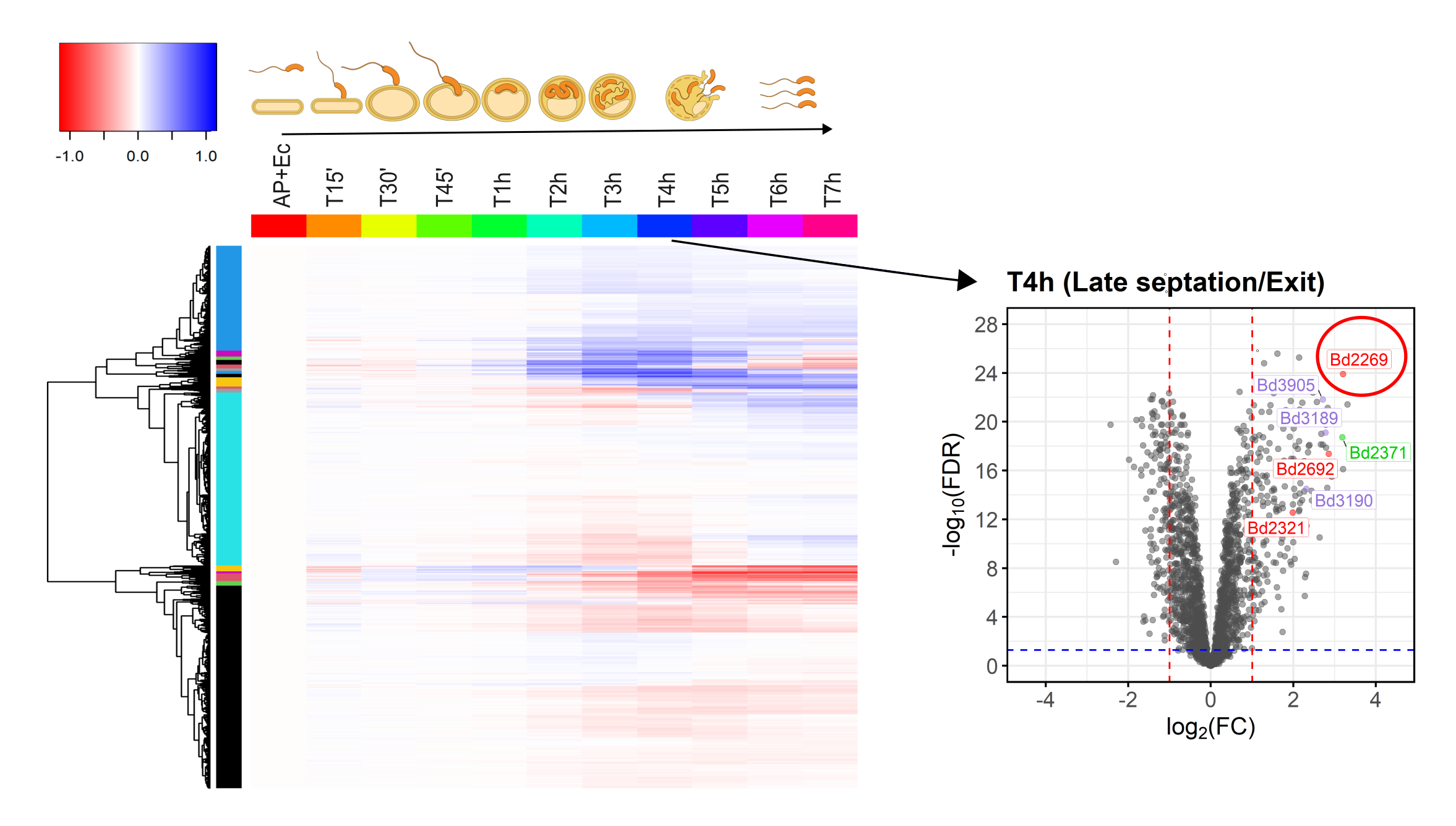
Fig.1. Overview of B. bacteriovorus proteins identified by quantitative proteomics over the whole predatory life cycle (T15’-T7h) on E. coli prey. A volcano plot from this data set identifies predatory protease Bd2269 up regulated in the prey exit phase (red encircled). In the heatmap the protein abundances were normalized to the ‘AP+Ec’ condition (starting at 0h, of free-swimming B. bacteriovorus only in ‘attack phase (AP)’ and ‘E. coli-only (Ec)’).
Video 1: Time-lapse microscopy of B. bacteriovorus exiting of prey cell remnants of Escherichia coli. (Video by S. Huwiler)
Towards the end of the life cycle when the predator exits prey remnants, we identified significant amounts of protease Bd2269. Gene knockout and heterologous expression experiments revealed that Bd2269 is involved in the prey exit process and damages E. coli from within. This quantitative predator proteome is a valuable resource to unravel bacterial predator-prey interactions and advances the search for novel antimicrobial enzymes.
Preprint available: https://doi.org/10.1101/2024.12.23.630089
People involved: Dr. Ting Lai (PhD), Denis Jankov (Msc), Dr. Simona Huwiler (PI)
Collaborators: Dr. Jonas Grossmann (Functional Genomics Centre Zurich), Dr. Bernd Roschitzki (Functional Genomics Centre Zurich)
Funding: The project 'Learning from predatory bacteria: from 'OMICS' to molecular mechanisms' is funded by a Swiss National Science Foundation (SNSF) Ambizione Fellowship. It is additionally supported by the Young Investigator Grant of Novartis Foundation for medical-biological Research and Gebauer Foundation.
Evolution of prey bacteria under predatory pressure
To combat antimicrobial resistant pathogens, natural predatory bacteria, like Bdellovibrio bacteriovorus, represent potential alternatives. B. bacteriovorus could be particularly potent as it kills a broad range of human bacterial pathogens, however, it remains unclear whether prey can evolve genetically-determined resistance against predation. We show that the model bacterium Escherichia coli K-12 consistently evolves resistance against B. bacteriovorus during experimental evolution. Selection for resistance scaled positively with predation pressure and was widespread after two cycles of predator exposure. Like antibiotics, predation resistance was costly, manifesting in a trade-off between predation resistance and fitness in the absence of predators. Genetic analysis identified changes in outer membrane porin OmpF as common resistance mechanism, while a mutation in cell envelope lipopolysaccharide-modifying enzyme WaaF was rarer but also conferred predation resistance. Our study uncovers evolutionary and mechanistic aspects of prey escape from predation, generating important knowledge on predator-prey interactions and to advance sustainable treatments.
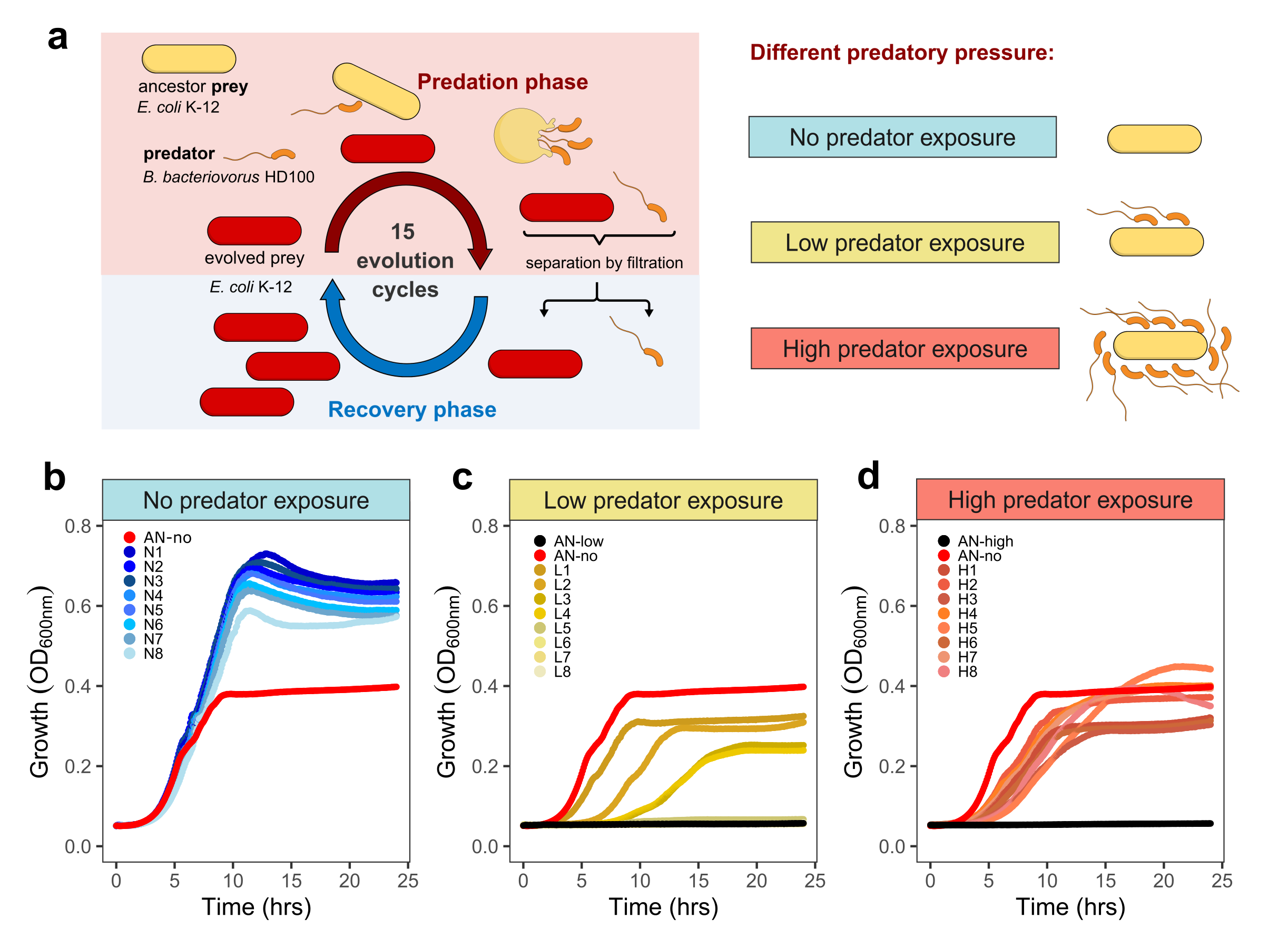
Fig.1. Experimental evolution experiment of E. coli prey bacteria repeatedly exposed to different predatory pressure by B. bacteriovorus. a. Experimental layout of evolution cycles changing from phases where E. coli prey is exposed to predation (predation phase) and not (recovery phase). Eight different prey lineages are exposed repeatedly to different concentration of the predator: no (N1-N8, blue), low (L1-L8, yellow) and high (H1-H8, red) predatory pressure. AN stands for the ancestral prey lineage. Growth dynamics of: b. prey lineages not exposed to predation, showing media adaptation. c. Low predatory pressure exposed prey lineages regain capability to grow under low predatory pressure. d. High predatory pressure exposed prey lineages regain capability to grow under high predatory pressure. In our study we show that there is a genetic bases associated with predation resistance, mostly due to changes in outer membrane porin F.
Preprint available: https://doi.org/10.1101/2024.09.27.615459
People involved: Dr. Subham Mridha (Postdoc, Co-Corresponding), Sarah-Luise Loose (Msc), Ove Ch. Mattmann (Msc, shared with Kümmerli lab), Saskia Isele (Msc), Dr. Simona Huwiler (PI)
Collaborator: Prof. Rolf Kümmerli (Department of Quantitative Biomedicine, University of Zurich)
Funding: The project is enabled by FAN (Research Talent Development Fund of the University of Zurich Alumni), the Ernst Göhner Stiftung, the Kurt und Senta Herrmann-Stiftung and an SNSF Ambizione grant.
Toolbox to modulate gene expression and protein secretion
The predatory bacterium Bdellovibrio bacteriovorus kills and consumes other bacteria, including dangerous pathogens. It lives in a variety of environments and has great potential in medicine, agriculture, and biotechnology. Due to its unique lifestyle, B. bacteriovorus holds great promise as an alternative to traditional antimicrobials against drug-resistant pathogens and in preventing food waste. Furthermore, predatory enzymes possess biotechnological potential. However, research and engineering efforts have been limited by the lack of genetic tools tailored to this organism. We developed a molecular toolbox to modulate gene expression and protein secretion in B. bacteriovorus. We tested a range of native and synthetic gene promoters at population and single-cell levels and evaluated the effect of different ribosomal binding sites. Further, we established a protocol to quantify extracellular release of a reporter protein. By enabling more precise genetic control, our work brings B. bacteriovorus a step closer to practical use as a biological tool to address antibiotic resistance and other microbial challenges.
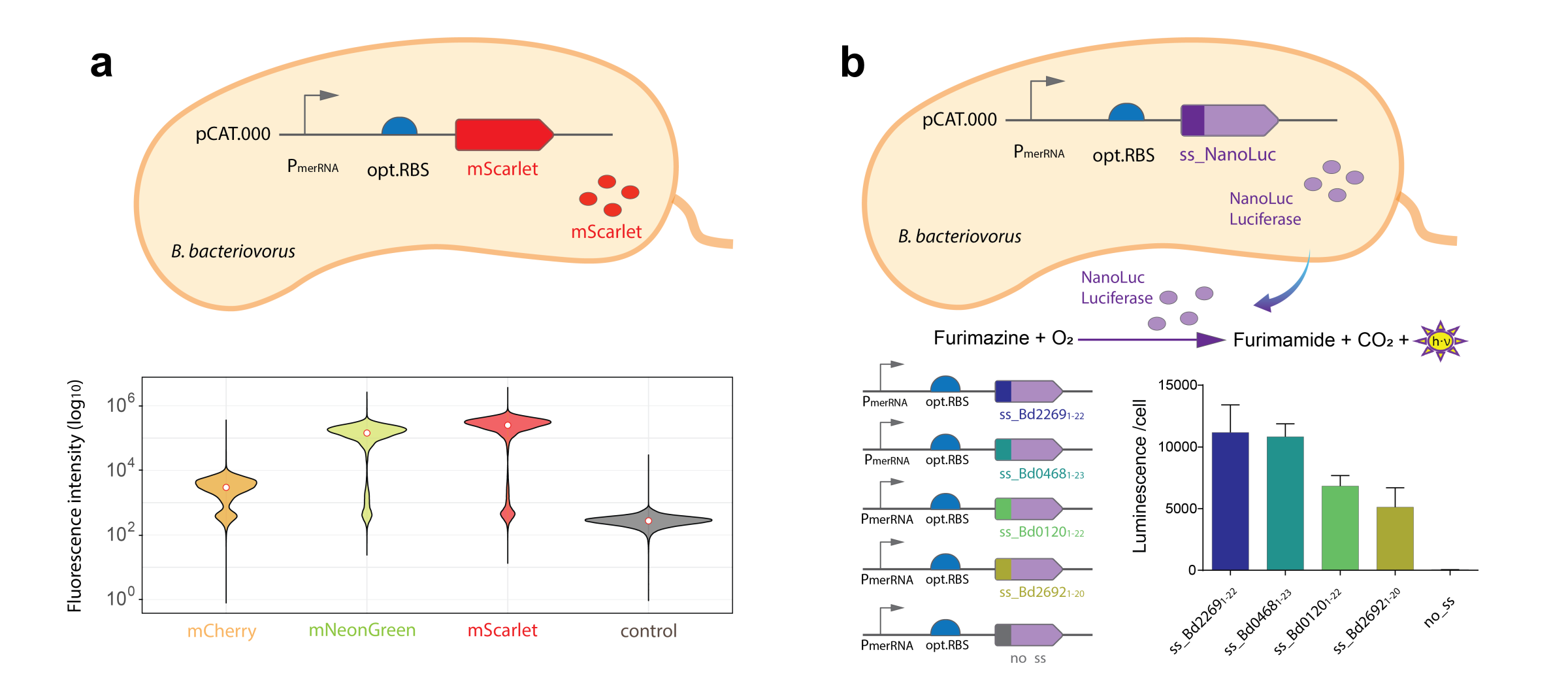
Fig.1. Comparison of different fluorescent proteins and secretion signals in B. bacteriovorus. a. Fluorescence intensity of the free-swimming B. bacteriovorus populations expressing different fluorescent proteins within, showing the best signal with mScarlet. b. Effect of different protein secretion signals on the secretion by B. bacteriovorus quantified with a NanoLuc Luciferase assay.
Preprint available: https://doi.org/10.1101/2025.05.17.654431
People involved: Dr. Ljiljana Mihajlovic (Postdoc), Lara M. Hofacker (Bsc), Florian Lindner (Postdoc), Dr. Simona Huwiler (PI)
Collaborators: Dr. Priyanikha Jayakumar (Flow cytometry Facility, University of Zurich), Prof. Andreas Diepold (Karlsruhe Institute of Technology, D)
Funding: The project is funded by the Vontobel-Stiftung, an UZH Innovation grant, an SNSF SPARK grant, a FreeNovation grant from Novartis Foundation, and an SNSF Ambizione grant.